Page 192 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioChemistry
P. 192
P1: GPB Final Pages
Encyclopedia of Physical Science and Technology EN013D-617 July 27, 2001 11:42
232 Protein Synthesis
binding fold now known to be typical of G-proteins. Do- Upon comparison with the EF-Tu ternary complex struc-
mains 2 and 3 are β-barrels, held together by strong inter- ture, EF-G was seen to have a conformation similar to that
domain contacts such that the two domains act as a single of the ternary complex, with Domains 3, 4, and 5 of EF-G
structural unit. Crystal structures of the many functional mimicking AA-tRNA.
forms of EF-Tu have been determined—EF-Tu:GDPNP The remarkable mimicry of tRNA by portions of
(a nonhydrolyzable analog of GTP), EF-Tu:GDP, and EF-G suggested a mechanism by which the factor might
complexes with EF-Ts or AA–tRNA. The orientation of be facilitating translocation. It is attractive to imagine
Domain1withrespecttoDomain2/3variesgreatlyamong that EF-G actively “chases” the peptidyl–tRNA into
the functional forms of EF-Tu. The two parts of the protein the ribosomal P-site because it mimics A-site-bound
◦
rotate by 90 when GDP–GTP exchange is catalyzed by tRNA. Furthermore, the sequence of events dictates that,
EF-Ts; this conformational change is due to two switch re- immediately after translocation, EF-G:GDP is released
gions in the G-domain. By altering small secondary struc- and another ternary complex (EF-Tu:GTP:AA–tRNA)
ture elements, these regions trigger long-range effects. enters the ribosomal A-site. Therefore the departure of
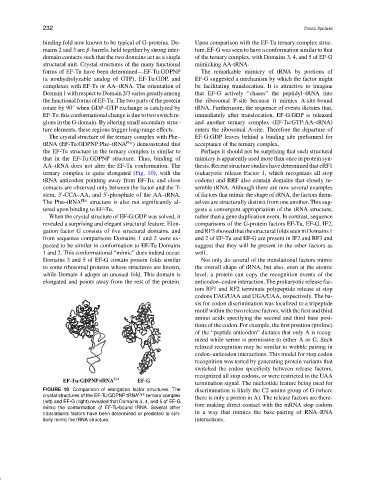
The crystal structure of the ternary complex with Phe– EF-G:GDP leaves behind a binding site preformed for
tRNA (EF-Tu:GDPNP:Phe–tRNA Phe ) demonstrated that acceptance of the ternary complex.
the EF-Tu structure in the ternary complex is similar to Perhaps it should not be surprising that such structural
that in the EF-Tu:GDPNP structure. Thus, binding of mimicry is apparently used more than once in protein syn-
AA–tRNA does not alter the EF-Tu conformation. The thesis. Recent structure studies have determined that eRF1
ternary complex is quite elongated (Fig. 10), with the (eukaryotic release Factor 1, which recognizes all stop
tRNA anticodon pointing away from EF-Tu, and close codons) and RRF also contain domains that closely re-
contacts are observed only between the factor and the T- semble tRNA. Although there are now several examples
stem, 3 -CCA–AA, and 5 -phosphate of the AA–tRNA. of factors that mimic the shape of tRNA, the factors them-
The Phe–tRNA Phe structure is also not significantly al- selves are structurally distinct from one another. This sug-
tered upon binding to EF-Tu. gests a convergent appropriation of the tRNA structure,
When the crystal structure of EF-G:GDP was solved, it rather than a gene duplication event. In contrast, sequence
revealed a surprising and elegant structural feature. Elon- comparisons of the G-protein factors EF-Tu, EF-G, IF2,
gation factor G consists of five structural domains, and andRF3showedthatthestructuralfoldsseeninDomains1
from sequence comparisons Domains 1 and 2 were ex- and 2 of EF-Tu and EF-G are present in IF2 and RF3 and
pected to be similar in conformation to EF-Tu Domains suggest that they will be present in the other factors as
1 and 2. This conformational “mimic” does indeed occur. well.
Domains 3 and 5 of EF-G contain protein folds similar Not only do several of the translational factors mimic
to some ribosomal proteins whose structures are known, the overall shape of tRNA, but also, even at the atomic
while Domain 4 adopts an unusual fold. This domain is level, a protein can copy the recognition events of the
elongated and points away from the rest of the protein. anticodon–codon interaction. The prokaryotic release fac-
tors RF1 and RF2 terminate polypeptide release at stop
codons UAG/UAA and UGA/UAA, respectively. The ba-
sis for codon discrimination was localized to a tripeptide
motif within the two release factors, with the first and third
amino acids specifying the second and third base posi-
tions of the codon. For example, the first position (proline)
of the “peptide anticodon” dictates that only A is recog-
nized while serine is permissive to either A or G. Such
relaxed recognition may be similar to wobble pairing in
codon–anticodon interactions. This model for stop codon
recognition was tested by generating protein variants that
switched the codon specificity between release factors,
recognized all stop codons, or were restricted to the UAA
termination signal. The nucleotide feature being used for
FIGURE 10 Comparison of elongation factor structures. The discrimination is likely the C2 amino group of G (where
crystal structures of the EF-Tu:GDPNP:tRNA Cys ternary complex there is only a proton in A). The release factors are there-
(left) and EF-G (right) revealed that Domains 3, 4, and 5 of EF-G
mimic the conformation of EF-Tu-bound tRNA. Several other fore making direct contact with the mRNA stop codons
translational factors have been determined or predicted to sim- in a way that mimics the base-pairing of RNA–RNA
ilarly mimic the tRNA structure. interactions.