Page 187 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioChemistry
P. 187
P1: GPB Final Pages
Encyclopedia of Physical Science and Technology EN013D-617 July 27, 2001 11:42
Protein Synthesis 227
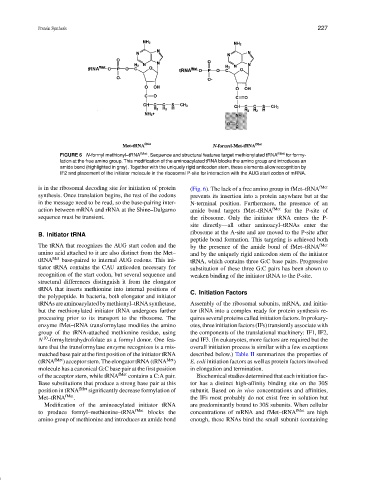
FIGURE 6 N-formyl methionyl–tRNA fMet . Sequence and structural features target methionylated tRNA fMet for formy-
lation at the free amino group. This modification of the aminoacylated tRNA blocks the amino group and introduces an
amide bond (highlighted in gray). Together with the uniquely rigid anticodon stem, these elements allow recognition by
IF2 and placement of the initiator molecule in the ribosomal P-site for interaction with the AUG start codon of mRNA.
is in the ribosomal decoding site for initiation of protein (Fig. 6). The lack of a free amino group in fMet–tRNA fMet
synthesis. Once translation begins, the rest of the codons prevents its insertion into a protein anywhere but at the
in the message need to be read, so the base-pairing inter- N-terminal position. Furthermore, the presence of an
action between mRNA and rRNA at the Shine–Dalgarno amide bond targets fMet–tRNA fMet for the P-site of
sequence must be transient. the ribosome. Only the initiator tRNA enters the P-
site directly—all other aminoacyl–tRNAs enter the
ribosome at the A-site and are moved to the P-site after
B. Initiator tRNA
peptide bond formation. This targeting is achieved both
The tRNA that recognizes the AUG start codon and the by the presence of the amide bond of fMet–tRNA fMet
amino acid attached to it are also distinct from the Met– and by the uniquely rigid anticodon stem of the initiator
tRNA Met base-paired to internal AUG codons. This ini- tRNA, which contains three G:C base pairs. Progressive
tiator tRNA contains the CAU anticodon necessary for substitution of these three G:C pairs has been shown to
recognition of the start codon, but several sequence and weaken binding of the initiator tRNA to the P-site.
structural differences distinguish it from the elongator
tRNA that inserts methionine into internal positions of
C. Initiation Factors
the polypeptide. In bacteria, both elongator and initiator
tRNAsareaminoacylatedbymethionyl–tRNAsynthetase, Assembly of the ribosomal subunits, mRNA, and initia-
but the methionylated initiator tRNA undergoes further tor tRNA into a complex ready for protein synthesis re-
processing prior to its transport to the ribosome. The quires several proteins called initiation factors. In prokary-
enzyme fMet–tRNA transformylase modifies the amino otes, three initiation factors (IFs) transiently associate with
group of the tRNA-attached methionine residue, using the components of the translational machinery: IF1, IF2,
10
N -formyltetrahydrofolate as a formyl donor. One fea- and IF3. (In eukaryotes, more factors are required but the
ture that the transformylase enzyme recognizes is a mis- overall initiation process is similar with a few exceptions
matched base pair at the first position of the initiator tRNA described below.) Table II summarizes the properties of
(tRNA fMet ) acceptor stem. The elongator tRNA (tRNA Met ) E. coli initiation factors as well as protein factors involved
molecule has a canonical G:C base pair at the first position in elongation and termination.
of the acceptor stem, while tRNA fMet contains a C:A pair. Biochemical studies determined that each initiation fac-
Base substitutions that produce a strong base pair at this tor has a distinct high-affinity binding site on the 30S
position in tRNA fMet significantly decrease formylation of subunit. Based on in vivo concentrations and affinities,
Met–tRNA fMet . the IFs most probably do not exist free in solution but
Modification of the aminoacylated initiator tRNA are predominantly bound to 30S subunits. When cellular
to produce formyl–methionine–tRNA fMet blocks the concentrations of mRNA and fMet–tRNA fMet are high
amino group of methionine and introduces an amide bond enough, these RNAs bind the small subunit (containing