Page 186 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioChemistry
P. 186
P1: GPB Final Pages
Encyclopedia of Physical Science and Technology EN013D-617 July 27, 2001 11:42
226 Protein Synthesis
IV. AN OVERVIEW OF TRANSLATION
Translation of an mRNA message into its polypeptide
product on the ribosome is a polymerization reaction,
and can be divided into three phases: initiation, elonga-
tion, and termination. Initiation requires the assembly of
the translational machinery from its individual compo-
nents to form a complex that is primed for peptide bond
formation. Formation of this initiation complex is usu-
ally the rate-limiting step in protein synthesis. Elonga-
tion is the sequential joining of amino acids by peptide
bonds as dictated by the codons of the mRNA. Trans-
lation proceeds along the mRNA in the 5 to 3 direc-
tion, and the resulting polypeptide chain is synthesized
in the amino-terminal to carboxy-terminal direction (N-
to C-terminal). Termination occurs when a stop codon is
reached in the message and the polypeptide is released
from the ribosome, which is then recycled for transla-
tion of another protein. Each step has associated protein
factors that facilitate substrate transport and ribosomal
function.
The ribosome is a ribonucleoprotein particle, that is,
it contains both RNA and protein components. The three
RNA molecules (in prokaryotes, four in eukaryotes) and
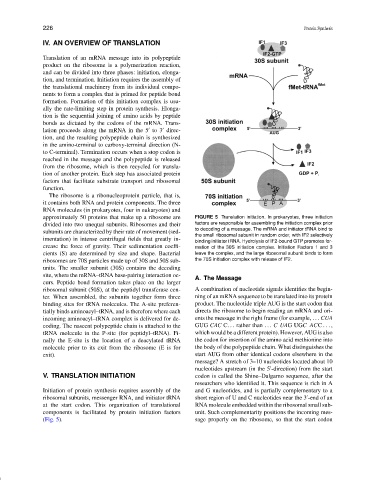
approximately 50 proteins that make up a ribosome are FIGURE 5 Translation initiation. In prokaryotes, three initiation
divided into two unequal subunits. Ribosomes and their factors are responsible for assembling the initiation complex prior
subunits are characterized by their rate of movement (sed- to decoding of a message. The mRNA and initiator tRNA bind to
the small ribosomal subunit in random order, with IF2 selectively
imentation) in intense centrifugal fields that greatly in-
binding initiator tRNA. Hydrolysis of IF2-bound GTP promotes for-
crease the force of gravity. Their sedimentation coeffi- mation of the 30S initiation complex. Initiation Factors 1 and 3
cients (S) are determined by size and shape. Bacterial leave the complex, and the large ribosomal subunit binds to form
ribosomes are 70S particles made up of 30S and 50S sub- the 70S initiation complex with release of IF2.
units. The smaller subunit (30S) contains the decoding
site, where the mRNA–tRNA base-pairing interaction oc-
A. The Message
curs. Peptide bond formation takes place on the larger
ribosomal subunit (50S), at the peptidyl transferase cen- A combination of nucleotide signals identifies the begin-
ter. When assembled, the subunits together form three ning of an mRNA sequence to be translated into its protein
binding sites for tRNA molecules. The A-site preferen- product. The nucleotide triple AUG is the start codon that
tially binds aminoacyl–tRNA, and is therefore where each directs the ribosome to begin reading an mRNA and ori-
incoming aminoacyl–tRNA complex is delivered for de- ents the message in the right frame (for example, ... CUA
coding. The nascent polypeptide chain is attached to the GUG CAC C... rather than ... CUAGUGCACC ... ,
tRNA molecule in the P-site (for peptidyl–tRNA). Fi- which would be a different protein). However, AUG is also
nally the E-site is the location of a deacylated tRNA the codon for insertion of the amino acid methionine into
molecule prior to its exit from the ribosome (E is for the body of the polypeptide chain. What distinguishes the
exit). start AUG from other identical codons elsewhere in the
message? A stretch of 3–10 nucleotides located about 10
nucleotides upstream (in the 5 -direction) from the start
V. TRANSLATION INITIATION codon is called the Shine–Dalgarno sequence, after the
researchers who identified it. This sequence is rich in A
Initiation of protein synthesis requires assembly of the and G nucleotides, and is partially complementary to a
ribosomal subunits, messenger RNA, and initiator tRNA short region of U and C nucleotides near the 3 -end of an
at the start codon. This organization of translational RNA molecule embedded within the ribosomal small sub-
components is facilitated by protein initiation factors unit. Such complementarity positions the incoming mes-
(Fig. 5). sage properly on the ribosome, so that the start codon