Page 181 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioChemistry
P. 181
P1: GPB Final Pages
Encyclopedia of Physical Science and Technology EN013D-617 July 27, 2001 11:42
Protein Synthesis 221
sources, and cofactors. Peptide bond formation takes TABLE I The Standard Genetic Code
place rapidly at the ribosome, with as many as 40 amino Second position
First Third
acids per second joined to a growing polypeptide chain.
position U C A G position
Yet coupled with the need for speed is the requirement
for accuracy. The misincorporation of a single amino acid U UUU Phe UCU Ser UAU Tyr UGU Cys U
could have drastic effects on the structure or function of a UUC Phe UCC Ser UAC Tyr UGC Cys C
newly synthesized protein. However, protein synthesis is UUA Leu UCA Ser UAA Stop UGA Stop A
accurate, with errors occurring on the average only once UUG Leu UCG Ser UAG Stop UGG Trp G
in 10,000 peptide bonds formed. C CUU Leu CCU Pro CAU His CGU Arg U
Formation of peptide bonds linking together amino CUC Leu CCC Pro CAC His CGC Arg C
acids could theoretically occur such that random se- CUA Leu CCA Pro CAA Gln CGA Arg A
quences are generated. Some of these sequences could CUG Leu CCG Pro CAG Gln CGG Arg G
result in a polypeptide that has a useful function. However, A AUU Ile ACU Thr AAU Asn AGU Ser U
transfer of genetic information from one generation to the AUC Ile ACC Thr AAC Asn AGC Ser C
next requires a systematic and reproducible mechanism AUA Ile ACA Thr AAA Lys AGA Arg A
for generating defined sequences. Polypeptide formation AUG Met a ACG Thr AAG Lys AGG Arg G
as we know it today is template-directed, with the mes- G GUU Val GCU Ala GAU Asp GGU Gly U
senger RNA (mRNA) copy of a gene providing the text to GUC Val GCC Ala GAC Asp GGC Gly C
be deciphered into the protein product. GUA Val GCA Ala GAA Glu GGA Gly A
Thesimplestlinkbetweennucleicacidandproteincom- GUG Val GCG Ala GAG Glu GGG Gly G
ponents would have been a code with a one-to-one cor- a
The AUG codon specifies the start of protein synthesis as well as
respondence where each nucleotide dictated a particular
internal methionine residues.
amino acid. With only four nucleotides making up the
information storage in cells, the resulting proteins synthe- II. TRANSFER RNAs
sized in such a scenario would be limited to those having
4 different amino acids. Even a code of two nucleotides Although an mRNA nucleotide sequence dictates the
per amino acid would allow for only 16 amino acids. The polypeptide sequence to be made, mRNAs do not directly
standard genetic code instead makes use of trinucleotide recognize amino acids. Amino acids are instead linked to
sequences called codons; these 64 codons are able to de- transfer RNA (tRNA) “adaptor” molecules, which serve
termine fully the 20 amino acids used in protein synthesis as reading heads to decipher the codons of mRNA through
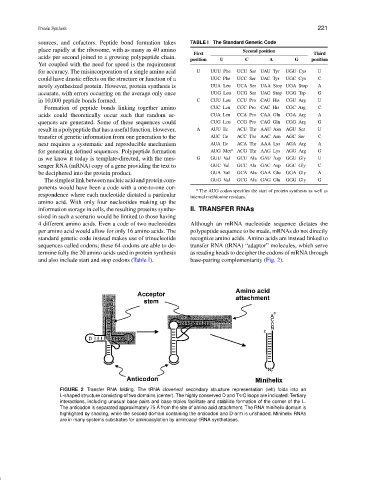
and also include start and stop codons (Table I). base-pairing complementarity (Fig. 2).
FIGURE 2 Transfer RNA folding. The tRNA cloverleaf secondary structure representation (left) folds into an
L-shaped structure consisting of two domains (center). The highly conserved D and T C loops are indicated. Tertiary
interactions, including unusual base pairs and base triples facilitate and stabilize formation of the corner of the L.
The anticodon is separated approximately 75 ˚ A from the site of amino acid attachment. The RNA minihelix domain is
highlighted by shading, while the second domain containing the anticodon and D-arm is unshaded. Minihelix RNAs
are in many systems substrates for aminoacylation by aminoacyl-tRNA synthetases.