Page 191 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioChemistry
P. 191
P1: GPB Final Pages
Encyclopedia of Physical Science and Technology EN013D-617 July 27, 2001 11:42
Protein Synthesis 231
be moved from the A-site to the P-site. The former
peptidyl–tRNA has been deacylated and needs to vacate
the P-site. Finally the mRNA must move three nucleotides
further in the 3 -direction so that the next codon can be
read. The concerted movement of tRNAs and mRNA at
the end of each elongation round is called translocation,
and is catalyzed by elongation factor G (EF-G), another
of the GTPase proteins in the translational machinery.
VII. TERMINATION
When the mRNA stop codon is reached, the fully synthe-
sized protein does not simply fall off the ribosome. Re-
lease factors (RFs) are the protein assistants that recognize
the presence of a stop codon in the ribosomal A-site and
trigger cleavage of the polypeptide from the P-site tRNA
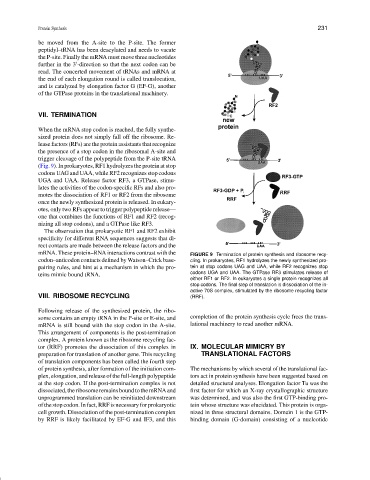
(Fig. 9). In prokaryotes, RF1 hydrolyzes the protein at stop
codons UAG and UAA, while RF2 recognizes stop codons
UGA and UAA. Release factor RF3, a GTPase, stimu-
lates the activities of the codon-specific RFs and also pro-
motes the dissociation of RF1 or RF2 from the ribosome
once the newly synthesized protein is released. In eukary-
otes, only two RFs appear to trigger polypeptide release—
one that combines the functions of RF1 and RF2 (recog-
nizing all stop codons), and a GTPase like RF3.
The observation that prokaryotic RF1 and RF2 exhibit
specificity for different RNA sequences suggests that di-
rect contacts are made between the release factors and the
mRNA. These protein–RNA interactions contrast with the FIGURE 9 Termination of protein synthesis and ribosome recy-
codon–anticodon contacts defined by Watson–Crick base- cling. In prokaryotes, RF1 hydrolyzes the newly synthesized pro-
pairing rules, and hint at a mechanism in which the pro- tein at stop codons UAG and UAA, while RF2 recognizes stop
teins mimic bound tRNA. codons UGA and UAA. The GTPase RF3 stimulates release of
either RF1 or RF2. In eukaryotes a single protein recognizes all
stop codons. The final step of translation is dissociation of the in-
active 70S complex, stimulated by the ribosome recycling factor
VIII. RIBOSOME RECYCLING (RRF).
Following release of the synthesized protein, the ribo-
some contains an empty tRNA in the P-site or E-site, and completion of the protein synthesis cycle frees the trans-
mRNA is still bound with the stop codon in the A-site. lational machinery to read another mRNA.
This arrangement of components is the post-termination
complex. A protein known as the ribosome recycling fac-
tor (RRF) promotes the dissociation of this complex in IX. MOLECULAR MIMICRY BY
preparation for translation of another gene. This recycling TRANSLATIONAL FACTORS
of translation components has been called the fourth step
of protein synthesis, after formation of the initiation com- The mechanisms by which several of the translational fac-
plex, elongation, and release of the full-length polypeptide tors act in protein synthesis have been suggested based on
at the stop codon. If the post-termination complex is not detailed structural analyses. Elongation factor Tu was the
dissociated, theribosome remains bound tothe mRNA and first factor for which an X-ray crystallographic structure
unprogrammed translation can be reinitiated downstream was determined, and was also the first GTP-binding pro-
of the stop codon. In fact, RRF is necessary for prokaryotic tein whose structure was elucidated. This protein is orga-
cell growth. Dissociation of the post-termination complex nized in three structural domains. Domain 1 is the GTP-
by RRF is likely facilitated by EF-G and IF3, and this binding domain (G-domain) consisting of a nucleotide