Page 29 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioChemistry
P. 29
P1: ZCK Final Pages
Encyclopedia of Physical Science and Technology EN005G-231 June 15, 2001 20:46
636 Enzyme Mechanisms
The substrates for most PLP-requiring processes are displaced lysine), with the pyridine ring of PLP acting
α-amino acids, and most of the processes take place at as an electron sink. For the racemases, a proton is
the α-carbon position, although some take place at the then delivered to the opposite face from the same or a
β-or γ -carbon. The enzymes which use PLP catalyze a different basic residue with the net result of inversion
wide range of reactions, including racemizations, decar- of configuration at the α-carbon. Attack of the active
boxylations, and amine transfers. In general, for all three site lysine effects product release and regenerates the
of these classes of reactions at the α-carbon the substrate cofactor.
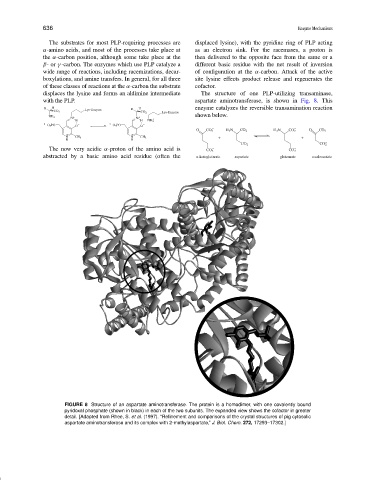
displaces the lysine and forms an aldimine intermediate The structure of one PLP-utilizing transaminase,
with the PLP. aspartate aminotransferase, is shown in Fig. 8. This
R H R H enzyme catalyzes the reversible transamination reaction
Lys-Enzyme
CO 2 CO 2 Lys-Enzyme
NH 2 N N shown below.
H H NH 2
2 O 3 PO O 2 O 3 PO O
O CO 2 H 3 N CO 2 H 3 N CO 2 O CO 2
N CH 3 N CH 3
H H
CO 2 CO 2
The now very acidic α-proton of the amino acid is CO 2 CO 2
abstracted by a basic amino acid residue (often the -ketoglutarate aspartate glutamate oxaloacetate
FIGURE 8 Structure of an aspartate aminotransferase. The protein is a homodimer, with one covalently bound
pyridoxal phosphate (shown in black) in each of the two subunits. The expanded view shows the cofactor in greater
detail. [Adapted from Rhee, S. et al. (1997). “Refinement and comparisons of the crystal structures of pig cytosolic
aspartate aminotransferase and its complex with 2-methylaspartate,” J. Biol. Chem. 272, 17293–17302.]