Page 25 - Academic Press Encyclopedia of Physical Science and Technology 3rd BioChemistry
P. 25
P1: ZCK Final Pages
Encyclopedia of Physical Science and Technology EN005G-231 June 15, 2001 20:46
632 Enzyme Mechanisms
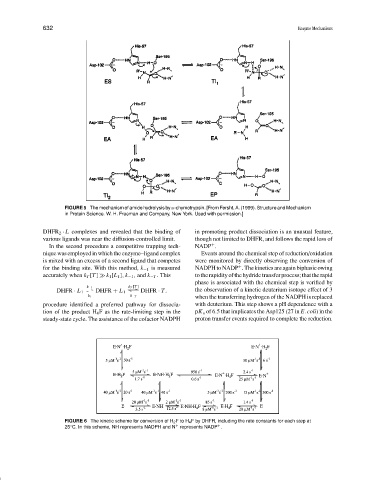
FIGURE 5 The mechanism of amide hydrolysis by α-chymotrypsin. [From Fersht, A. (1999). Structure and Mechanism
in Protein Science. W. H. Freeman and Company, New York. Used with permission.]
DHFR 2 · L complexes and revealed that the binding of in promoting product dissociation is an unusual feature,
various ligands was near the diffusion-controlled limit. though not limited to DHFR, and follows the rapid loss of
In the second procedure a competitive trapping tech- NADP .
+
nique was employed in which the enzyme–ligand complex Events around the chemical step of reduction/oxidation
is mixed with an excess of a second ligand that competes were monitored by directly observing the conversion of
for the binding site. With this method, k −1 is measured NADPH to NADP . The kinetics are again biphasic owing
+
accurately when k T [T ] k 1 [L 1 ], k −1 , and k −T . This totherapidityofthehydridetransferprocess;thattherapid
phase is associated with the chemical step is verified by
k T [T ]
k −1
DHFR · L 1 DHFR + L 1 DHFR · T. the observation of a kinetic deuterium isotope effect of 3
when the transferring hydrogen of the NADPH is replaced
k 1 k −T
procedure identified a preferred pathway for dissocia- with deuterium. This step shows a pH dependence with a
tion of the product H 4 F as the rate-limiting step in the pK a of 6.5 that implicates the Asp125 (27 in E. coli) in the
steady-state cycle. The assistance of the cofactor NADPH proton transfer events required to complete the reduction.
FIGURE 6 The kinetic scheme for conversion of H 2 FtoH 4 F by DHFR, including the rate constants for each step at
+
25 C. In this scheme, NH represents NADPH and N represents NADP .
◦
+