Page 147 - Introduction to Paleobiology and The Fossil Record
P. 147
134 INTRODUCTION TO PALEOBIOLOGY AND THE FOSSIL RECORD
lapsed like a pack of cards soon after. The “dinosaur” DNA turned out to be human: the PCR
technique is so sensitive that the tiniest fragment of DNA, in this case from sweat or sneezed mucus
of a lab assistant, can be amplified. Careful study showed that DNA is highly labile and breaks down
in even hundreds of years, and is pretty well all gone by 40,000 years, even in the most exceptional
preservation.
The most convincing studies of the DNA of fossil species come from ice age mammals such as
the cave bear, giant Irish deer, Neandertal man, woolly rhino and woolly mammoth. In a rush of
enthusiasm, three labs independently sequenced and published the complete mitochondrial genome
of the woolly mammoth in early 2006 (Krause et al. 2006; Poinar et al. 2006; Rogaev et al. 2006).
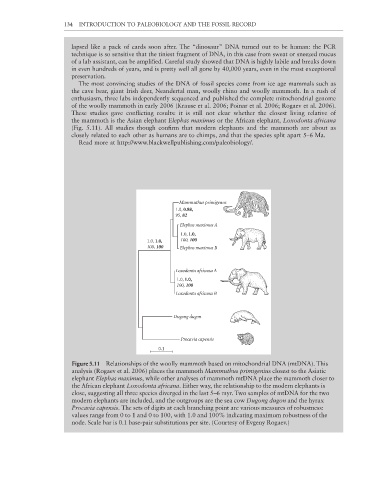
These studies gave conflicting results: it is still not clear whether the closest living relative of
the mammoth is the Asian elephant Elephas maximus or the African elephant, Loxodonta africana
(Fig. 5.11). All studies though confirm that modern elephants and the mammoth are about as
closely related to each other as humans are to chimps, and that the species split apart 5–6 Ma.
Read more at http://www.blackwellpublishing.com/paleobiology/.
Mammuthus primigenus
1.0, 0.88,
95, 82
Elephas maximus A
1.0, 1.0,
1.0, 1.0, 100, 100
100, 100 Elephas maximus B
Loxodonta africana A
1.0, 1.0,
100, 100
Loxodonta africana B
Dugong dugon
Procavia capensis
0.1
Figure 5.11 Relationships of the woolly mammoth based on mitochondrial DNA (mtDNA). This
analysis (Rogaev et al. 2006) places the mammoth Mammuthus primigenius closest to the Asiatic
elephant Elephas maximus, while other analyses of mammoth mtDNA place the mammoth closer to
the African elephant Loxodonta africana. Either way, the relationship to the modern elephants is
close, suggesting all three species diverged in the last 5–6 myr. Two samples of mtDNA for the two
modern elephants are included, and the outgroups are the sea cow Dugong dugon and the hyrax
Procavia capensis. The sets of digits at each branching point are various measures of robustness:
values range from 0 to 1 and 0 to 100, with 1.0 and 100% indicating maximum robustness of the
node. Scale bar is 0.1 base-pair substitutions per site. (Courtesy of Evgeny Rogaev.)