Page 135 - Thermodynamics of Biochemical Reactions
P. 135
132 Chapter 7 Thermodynamics of the Binding of Ligands by Proteins
-- [heme]
a
2x10-R 4x10 ' 6x 10" 8x10-R 1x10
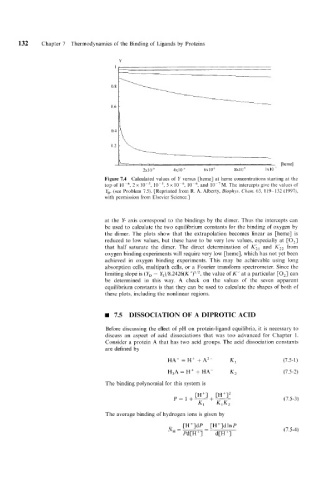
Figure 7.4 Calculated values of Y versus [heme] at heme concentrations starting at the
top of 2 x 5 x and 10-7M. The intercepts give the values of
Y,. (see Problem 7.5). [Reprinted from R. A. Alberty, Biopkys. Chem. 63, 119-132 (1997),
with permission from Elsevier Science.]
at the Y- axis correspond to the bindings by the dimer. Thus the intercepts can
be used to calculate the two equilibrium constants for the binding of oxygen by
the dimer. The plots show that the extrapolation becomes linear as [heme] is
reduced to low values, but these have to be very low values, especially at [O,]
that half saturate the dimer. The direct determination of KL1 and K;, from
oxygen binding experiments will require very low [heme], which has not yet been
achieved in oxygen binding experiments. This may be achievable using long
absorption cells, multipath cells, or a Fourier transform spectrometer. Since the
limiting slope is (Y,, - YT)/8.2426(K ")Ii2, the value of K" at a particular [O,] can
be determined in this way. A check on the values of the seven apparent
equilibrium constants is that they can be used to calculate the shapes of both of
these plots, including the nonlinear regions.
7.5 DISSOCIATION OF A DIPROTIC ACID
Before discussing the effect of pH on protein-ligand equilibria, it is necessary to
discuss an aspect of acid dissociations that was too advanced for Chapter 1.
Consider a protein A that has two acid groups. The acid dissociation constants
are defined by
HA- = H+ + A'- K, (7.5-1)
H,A = Hf + HA- K2 (7.5-2)
The binding polynomial for this system is
(7.5-3)
The average binding of hydrogen ions is given by
[H+]dP - [H+]dln P
..- (7.5-4)
" - Pd[H+] - d[Hf]