Page 243 - Thermodynamics of Biochemical Reactions
P. 243
Apparent Equilibrium Constants 243
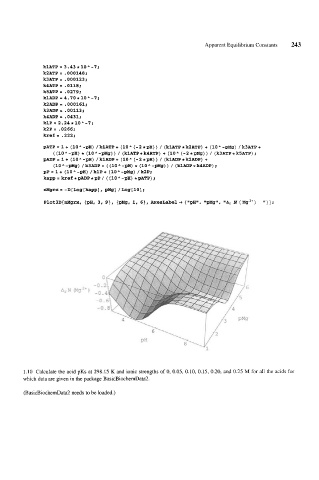
klATP = 3 -43 * 10 " -7;
k2ATP= .000148;
k3ATP= .000123;
k4ATP= -0118;
k5ATP = .0279;
klADP=4.70*10A-1;
k2ADP= .000161;
k3ADP= .00113;
k4ADP= .0431;
klP= 2.24*10"-7;
k2P= .0266;
kre€= -222;
pATP= 1+ (lOA-pH) /klATP+ (10" (-2*pH)) / (klATP*kSATP) + (lOA-pMg) /k3ATP+
((lOA-gH) * (lOA-gMg)) / (klATP*k4ATP) + (10" (-2*gMg)) / (k3ATP*kSATP);
pADP= 1+ (lOA-gH) /klADP+ (10" (-2*pH)) / (klADP*kZADP) +
(1O"-gMg) /k3ADP+ ((10"-pH) * (lOA-pMg)) / (klADP*k4ADP);
pP = 1 + (10 " -pH) / klP + (10 " -gMg) / k2P;
kapp = kref * PADP * pP / ( (10 " -BH) * pATP) ;
nMgrx= -D[Log[kapp], pMg] /Log[lo];
Plot3D[nMgrx, {gH, 3, g}, {pMg, 1, 61, AxesLabel -) {llgH1m, "pMg", "Az N (Mg*') "}I;
1.10 Calculate the acid pKs at 298.15 K and ionic strengths of 0, 0.05, 0.10, 0.15, 0.20, and 0.25 M for all the acids for
which data are given in the package BasicBiochemData2.
(BasicBiochemData2 needs to be loaded.)