Page 88 - Chiral Separation Techniques
P. 88
64 3 Combinatorial Approaches to Recognition of Chirality: Preparation …
fraction 1 fraction 2 fraction 3
80
Absorbance at 226 nm (–) 25 Percentage of Acetonitrile (- - - -)
0
0 10 20 30 40 50
Time (min)
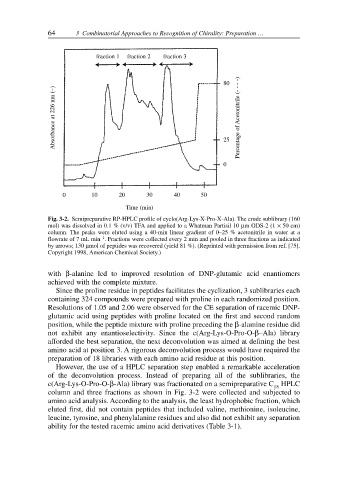
Fig. 3-2. Semipreparative RP-HPLC profile of cyclo(Arg-Lys-X-Pro-X-Ala). The crude sublibrary (160
mol) was dissolved in 0.1 % (v/v) TFA and applied to a Whatman Partisil 10 µm ODS-2 (1 × 50 cm)
column. The peaks were eluted using a 40-min linear gradient of 0–25 % acetonitrile in water at a
–1
flowrate of 7 mL min . Fractions were collected every 2 min and pooled in three fractions as indicated
by arrows; 130 µmol of peptides was recovered (yield 81 %). (Reprinted with permission from ref. [75].
Copyright 1998, American Chemical Society.)
with β-alanine led to improved resolution of DNP-glutamic acid enantiomers
achieved with the complete mixture.
Since the proline residue in peptides facilitates the cyclization, 3 sublibraries each
containing 324 compounds were prepared with proline in each randomized position.
Resolutions of 1.05 and 2.06 were observed for the CE separation of racemic DNP-
glutamic acid using peptides with proline located on the first and second random
position, while the peptide mixture with proline preceding the β-alamine residue did
not exhibit any enantioselectivity. Since the c(Arg-Lys-O-Pro-O-β–Ala) library
afforded the best separation, the next deconvolution was aimed at defining the best
amino acid at position 3. A rigorous deconvolution process would have required the
preparation of 18 libraries with each amino acid residue at this position.
However, the use of a HPLC separation step enabled a remarkable acceleration
of the deconvolution process. Instead of preparing all of the sublibraries, the
c(Arg-Lys-O-Pro-O-β-Ala) library was fractionated on a semipreparative C HPLC
18
column and three fractions as shown in Fig. 3-2 were collected and subjected to
amino acid analysis. According to the analysis, the least hydrophobic fraction, which
eluted first, did not contain peptides that included valine, methionine, isoleucine,
leucine, tyrosine, and phenylalanine residues and also did not exhibit any separation
ability for the tested racemic amino acid derivatives (Table 3-1).