Page 136 - Computational Retinal Image Analysis
P. 136
5 Deep learning based methods 129
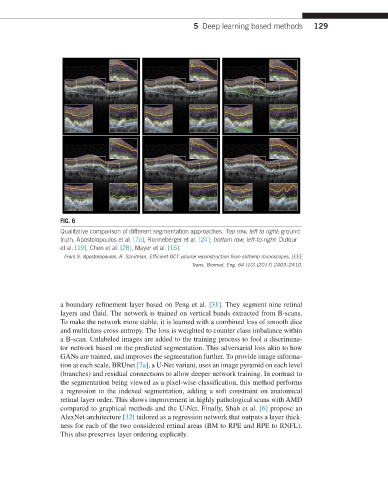
FIG. 6
Qualitative comparison of different segmentation approaches. Top row, left to right: ground
truth, Apostolopoulos et al. [7a], Ronneberger et al. [24]; bottom row, left-to-right: Dufour
et al. [19], Chen et al. [28], Mayer et al. [16].
From S. Apostolopoulos, R. Sznitman, Efficient OCT volume reconstruction from slitlamp microscopes, IEEE
Trans. Biomed. Eng. 64 (10) (2017) 2403–2410.
a boundary refinement layer based on Peng et al. [31]. They segment nine retinal
layers and fluid. The network is trained on vertical bands extracted from B-scans.
To make the network more stable, it is learned with a combined loss of smooth dice
and multiclass cross-entropy. The loss is weighted to counter class imbalance within
a B-scan. Unlabeled images are added to the training process to fool a discrimina-
tor network based on the predicted segmentation. This adversarial loss akin to how
GANs are trained, and improves the segmentation further. To provide image informa-
tion at each scale, BRUnet [7a], a U-Net variant, uses an image pyramid on each level
(branches) and residual connections to allow deeper network training. In contrast to
the segmentation being viewed as a pixel-wise classification, this method performs
a regression to the indexed segmentation, adding a soft constraint on anatomical
retinal layer order. This shows improvement in highly pathological scans with AMD
compared to graphical methods and the U-Net. Finally, Shah et al. [6] propose an
AlexNet-architecture [32] tailored as a regression network that outputs a layer thick-
ness for each of the two considered retinal areas (BM to RPE and RPE to RNFL).
This also preserves layer ordering explicitly.