Page 288 - Thermodynamics of Biochemical Reactions
P. 288
288 Mathematica Solutions to Problems
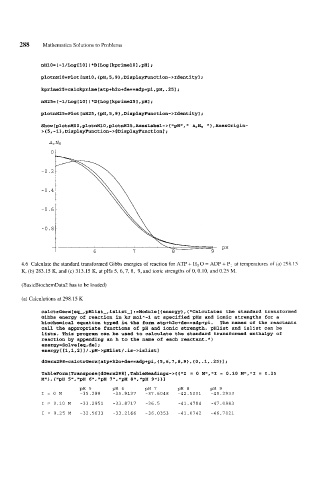
nH10= (-1/Log 1101 ) *D [Log IkprimelOI ,pH];
plotnH10=Plot[nH10,{~H,5,9},Disp~ayFunction-~Identityl;
kprime25=calckprime[atp+h20+de==adp+pi,pH,.251;
nH25=(-1/Log[101 )*D[Loglkprime251 ,PHI;
plotnH25=Plot[nH25,{~H,5,9},D~splayFunct~on->Ident~tyl;
Show [plotnHOO,plotnH1O,plotnH25 ,AxesLabel-> Cm'pKtl, ArNH I' 1, AxesOrigin-
>(5,-1},DisplayFunction->$Disp~ayFunction];
a, NH
ot
4.6 Calculate the standard transformed Gibbs energies of reaction for ATP + HzO = ADP + P, at temperatures of (a) 298.15
K, (b) 283.15 K, and (c) 313.15 K, at pHs 5, 6, 7, 8, 9, and ionic strengths of 0, 0.10, and 0.25 M.
(BasicBiochemData2 has to be loaded)
(a) Calculations at 298.15 K
calctrGerx[e~,pHlist_ri8lislist~]:=Module[{energy},(*Calculates the standard transformed
Gibbs energy of reaction in kJ molA-l at specified ~HS and ionic strengths for a
biochemical equation typed in the form atp+hao+de==adp+pi. The names of the reactants
call the appropriate functions of pH and ionic strength. pHlist and islist can be
lists. This program can be used to calculate the standard transformed enthalgy of
reaction by appending an h to the name of each reactant.*)
energy=Solve[eq,del;
energy[[1,1,2]l/.pH->~H1ist/.is-~islist]
dGerx298=calctrGerx[atp+h2o+de==adp+pi,{5,6,7,8,9},~0,.1,.25}1;
TableForm[Transpose[dGerx298],TableHeadings-> {"I =. 0 M","I = 0.10 M","I = 0.25
M"}, ("pH 5","pH 6","pH 7","pH 8","pH 9"}}1
PH 5 PH 6 PH 7 PH 8 PH 9
I=OM -35.299 -35.9137 -37.6048 -42.5001 -48.2933
I = 0.10 M -33.2951 -33.8717 -36.5 -41.4784 -47.0983
I z 0.25 M -32.5633 -33.2166 -36.0353 -41.0742 -46.7021