Page 176 - Advances in Biomechanics and Tissue Regeneration
P. 176
172 8. TOWARDS THE REAL-TIME MODELING OF THE HEART
Displacement
1.680e+01
12.727
8.6543
4.5815
5.087e-01
(A) (B)
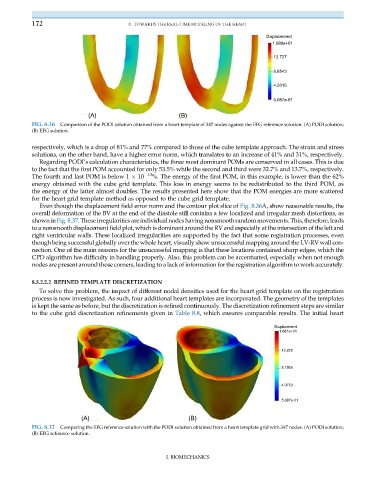
FIG. 8.36 Comparison of the PODI solution obtained from a heart template of 347 nodes against the EFG reference solution. (A) PODI solution;
(B) EFG solution.
respectively, which is a drop of 81% and 77% compared to those of the cube template approach. The strain and stress
solutions, on the other hand, have a higher error norm, which translates to an increase of 41% and 31%, respectively.
Regarding PODI’s calculation characteristics, the three most dominant POMs are conserved in all cases. This is due
to the fact that the first POM accounted for only 53.5% while the second and third were 32.7% and 13.7%, respectively.
The fourth and last POM is below 1 10 13 %. The energy of the first POM, in this example, is lower than the 62%
energy obtained with the cube grid template. This loss in energy seems to be redistributed to the third POM, as
the energy of the latter almost doubles. The results presented here show that the POM energies are more scattered
for the heart grid template method as opposed to the cube grid template.
Even though the displacement field error norm and the contour plot slice of Fig. 8.36A, show reasonable results, the
overall deformation of the BV at the end of the diastole still contains a few localized and irregular mesh distortions, as
showninFig. 8.37. Theseirregularities are individual nodes havingnonsmoothrandommovements.This,therefore,leads
to a nonsmooth displacement field plot, which is dominant around the RV and especially at the intersection of the left and
right ventricular walls. These localized irregularities are supported by the fact that some registration processes, even
though being successful globally over the whole heart, visually show unsuccessful mapping around the LV-RV wall con-
nection. One of the main reasons for the unsuccessful mapping is that these locations contained sharp edges, which the
CPD algorithm has difficulty in handling properly. Also, this problem can be accentuated, especially when not enough
nodes are present around those corners, leading to a lack of information for the registration algorithm to work accurately.
8.5.2.2.2 REFINED TEMPLATE DISCRETIZATION
To solve this problem, the impact of different nodal densities used for the heart grid template on the registration
process is now investigated. As such, four additional heart templates are incorporated. The geometry of the templates
is kept the same as before, but the discretization is refined continuously. The discretization refinement steps are similar
to the cube grid discretization refinements given in Table 8.8, which ensures comparable results. The initial heart
Displacement
1.681e+01
12.226
8.1506
4.0753
5.087e-01
(A) (B)
FIG. 8.37 Comparing the EFG reference solution with the PODI solution obtained from a heart template grid with 347 nodes. (A) PODI solution;
(B) EFG reference solution.
I. BIOMECHANICS