Page 266 - Big Data Analytics for Intelligent Healthcare Management
P. 266
10.7 MOLECULAR INTERACTIONS AND SCORING FUNCTIONS 259
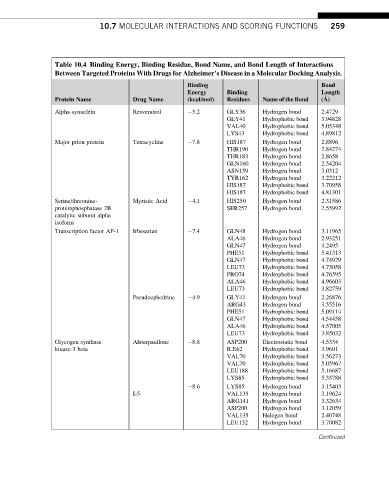
Table 10.4 Binding Energy, Binding Residue, Bond Name, and Bond Length of Interactions
Between Targeted Proteins With Drugs for Alzheimer’s Disease in a Molecular Docking Analysis.
Binding Bond
Energy Binding Length
˚
Protein Name Drug Name (kcal/mol) Residues Name of the Bond (A)
Alpha-synuclein Resveratrol 5.2 GLY36 Hydrogen bond 2.4729
GLY41 Hydrophobic bond 3.94828
VAL40 Hydrophobic bond 5.05348
LYS43 Hydrophobic bond 4.89812
Major prion protein Tetracycline 7.8 HIS187 Hydrogen bond 2.8896
THR190 Hydrogen bond 2.84274
THR183 Hydrogen bond 2.8658
GLN160 Hydrogen bond 2.54204
ASN159 Hydrogen bond 3.0312
TYR162 Hydrogen bond 3.22212
HIS187 Hydrophobic bond 3.70958
HIS187 Hydrophobic bond 4.81301
Serine/threonine- Myristic Acid 4.1 HIS250 Hydrogen bond 2.31586
proteinphosphatase 2B SER257 Hydrogen bond 2.55992
catalytic subunit alpha
isoform
Transcription factor AP-1 Irbesartan 7.4 GLN48 Hydrogen bond 3.11965
ALA46 Hydrogen bond 2.93251
GLN47 Hydrogen bond 3.2495
PHE51 Hydrophobic bond 5.41313
GLN47 Hydrophobic bond 4.74929
LEU73 Hydrophobic bond 4.75058
PRO74 Hydrophobic bond 4.76595
ALA46 Hydrophobic bond 4.96603
LEU73 Hydrophobic bond 3.82759
Pseudoephedrine 4.9 GLY41 Hydrogen bond 2.26876
ARG43 Hydrogen bond 3.55516
PHE51 Hydrophobic bond 5.09114
GLN47 Hydrophobic bond 4.54458
ALA46 Hydrophobic bond 4.57005
LEU73 Hydrophobic bond 3.85032
Glycogen synthase Alsterpaullone 8.8 ASP200 Electrostatic bond 4.5334
kinase-3 beta ILE62 Hydrophobic bond 3.9601
VAL70 Hydrophobic bond 3.56273
VAL70 Hydrophobic bond 5.05967
LEU188 Hydrophobic bond 5.16687
LYS85 Hydrophobic bond 5.35788
8.6 LYS85 Hydrogen bond 3.15403
I-5 VAL135 Hydrogen bond 3.19624
ARG141 Hydrogen bond 3.32634
ASP200 Hydrogen bond 3.12059
VAL135 Halogen bond 2.40748
LEU132 Hydrogen bond 3.70082
Continued