Page 127 - Cascade_Biocatalysis_Integrating_Stereoselective_and_Environmentally_Friendly_Reactions
P. 127
5.2 Physiological Cascade Reactions Involving P450s 103
O
S N O
N
HN
Omeprazole
CYP2C19 CYP3A4
OH
5 O
O
S N O S N O
N N
O
HN HN
5-Hydroxyomeprazole Omeprazole sulfone
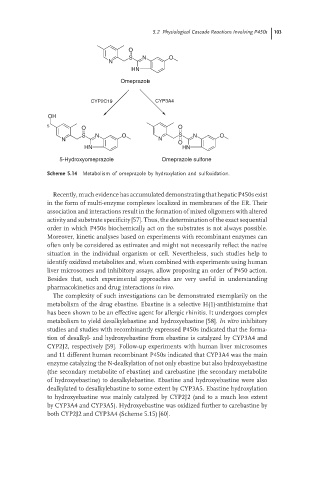
Scheme 5.14 Metabolism of omeprazole by hydroxylation and sulfoxidation.
Recently, much evidence has accumulated demonstrating that hepatic P450s exist
in the form of multi-enzyme complexes localized in membranes of the ER. Their
association and interactions result in the formation of mixed oligomers with altered
activity and substrate specificity [57]. Thus, the determination of the exact sequential
order in which P450s biochemically act on the substrates is not always possible.
Moreover, kinetic analyses based on experiments with recombinant enzymes can
often only be considered as estimates and might not necessarily reflect the native
situation in the individual organism or cell. Nevertheless, such studies help to
identify oxidized metabolites and, when combined with experiments using human
liver microsomes and inhibitory assays, allow proposing an order of P450 action.
Besides that, such experimental approaches are very useful in understanding
pharmacokinetics and drug interactions in vivo.
The complexity of such investigations can be demonstrated exemplarily on the
metabolism of the drug ebastine. Ebastine is a selective H(1)-antihistamine that
has been shown to be an effective agent for allergic rhinitis. It undergoes complex
metabolism to yield desalkylebastine and hydroxyebastine [58]. In vitro inhibitory
studies and studies with recombinantly expressed P450s indicated that the forma-
tion of desalkyl- and hydroxyebastine from ebastine is catalyzed by CYP3A4 and
CYP2J2, respectively [59]. Follow-up experiments with human liver microsomes
and 11 different human recombinant P450s indicated that CYP3A4 was the main
enzyme catalyzing the N-dealkylation of not only ebastine but also hydroxyebastine
(the secondary metabolite of ebastine) and carebastine (the secondary metabolite
of hydroxyebastine) to desalkylebastine. Ebastine and hydroxyebastine were also
dealkylated to desalkylebastine to some extent by CYP3A5. Ebastine hydroxylation
to hydroxyebastine was mainly catalyzed by CYP2J2 (and to a much less extent
by CYP3A4 and CYP3A5). Hydroxyebastine was oxidized further to carebastine by
both CYP2J2 and CYP3A4 (Scheme 5.15) [60].