Page 310 - Advances in Biomechanics and Tissue Regeneration
P. 310
308 15. COMPUTATIONAL SIMULATION OF CELL BEHAVIOR FOR TISSUE REGENERATION
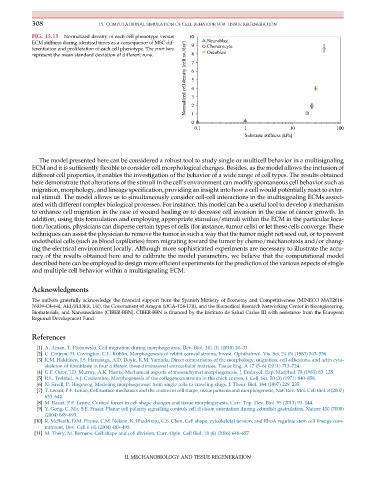
FIG. 15.15 Normalized density of each cell phenotype versus 10
ECM stiffness during identical times as a consequence of MSC dif- 9 Neuroblast
Normalized cell density [cell no./day] 6 5 4 3
ferentiation and proliferation of each cell phenotype. The error bars Chondrocyte
represent the mean standard deviation of different runs. 8 7 Oseoblast
0 2 1
0.1 1 10 100
Substrate stiffness [kPa]
The model presented here can be considered a robust tool to study single or multicell behavior in a multisignaling
ECM and it is sufficiently flexible to consider cell morphological changes. Besides, as the model allows the inclusion of
different cell properties, it enables the investigation of the behavior of a wide range of cell types. The results obtained
here demonstrate that alterations of the stimuli in the cell’s environment can modify spontaneous cell behavior such as
migration, morphology, and lineage specification, providing an insight into how a cell would potentially react to exter-
nal stimuli. The model allows us to simultaneously consider cell-cell interactions in the multisignaling ECMs associ-
ated with different complex biological processes. For instance, this model can be a useful tool to develop a mechanism
to enhance cell migration in the case of wound healing or to decrease cell invasion in the case of cancer growth. In
addition, using this formulation and employing appropriate stimulus/stimuli within the ECM in the particular loca-
tion/locations, physicians can disperse certain types of cells (for instance, tumor cells) or let these cells converge. These
techniques can assist the physician to remove the tumor in such a way that the tumor might not seed out, or to prevent
endothelial cells (such as blood capillaries) from migrating toward the tumor by chemo/mechanotaxis and/or chang-
ing the electrical environment locally. Although more sophisticated experiments are necessary to illustrate the accu-
racy of the results obtained here and to calibrate the model parameters, we believe that the computational model
described here can be employed to design more efficient experiments for the prediction of the various aspects of single
and multiple cell behavior within a multisignaling ECM.
Acknowledgments
The authors gratefully acknowledge the financial support from the Spanish Ministry of Economy and Competitiveness (MINECO MAT2016-
76039-C4-4-R, AEI/FEDER, UE), the Government of Aragon (DGA-T24-17R), and the Biomedical Research Networking Center in Bioengineering,
Biomaterials, and Nanomedicine (CIBER-BBN). CIBER-BBN is financed by the Instituto de Salud Carlos III with assistance from the European
Regional Development Fund.
References
[1] A. Aman, T. Piotrowski, Cell migration during morphogenesis, Dev. Biol. 341 (1) (2010) 20–33.
[2] C. Cintron, H. Covington, C.L. Kublin, Morphogenesis of rabbit corneal stroma, Invest. Ophthalmol. Vis. Sci. 24 (5) (1983) 543–556.
[3] K.M. Hakkinen, J.S. Harunaga, A.D. Doyle, K.M. Yamada, Direct comparisons of the morphology, migration, cell adhesions, and actin cyto-
skeleton of fibroblasts in four different three-dimensional extracellular matrices, Tissue Eng. A 17 (5–6) (2011) 713–724.
[4] G.F. Oster, J.D. Murray, A.K. Harris, Mechanical aspects of mesenchymal morphogenesis, J. Embryol. Exp. Morphol. 78 (1983) 83–125.
[5] R.L. Trelstad, A.J. Coulombre, Morphogenesis of the collagenous stroma in the chick cornea, J. Cell. Sci. 50 (3) (1971) 840–858.
[6] N. Savill, P. Hogeweg, Modeling morphogenesis: from single cells to crawling slugs, J. Theor. Biol. 184 (1997) 229–235.
[7] T. Lecuit, P.F. Lenne, Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis, Nat. Rev. Mol. Cell Biol. 8 (2007)
633–644.
[8] M. Rauzi, P.F. Lenne, Cortical forces in cell shape changes and tissue morphogenesis, Curr. Top. Dev. Biol. 95 (2011) 93–144.
[9] Y. Gong, C. Mo, S.E. Fraser, Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation, Nature 430 (7000)
(2004) 689–693.
[10] R. McBeath, D.M. Pirone, C.M. Nelson, K. Bhadriraju, C.S. Chen, Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage com-
mitment, Dev. Cell 6 (4) (2004) 483–495.
[11] M. Th ery, M. Bornens, Cell shape and cell division, Curr. Opin. Cell Biol. 18 (6) (2006) 648–657.
II. MECHANOBIOLOGY AND TISSUE REGENERATION