Page 75 - Artificial Intelligence for Computational Modeling of the Heart
P. 75
Chapter 2 Implementation of a patient-specific cardiac model 45
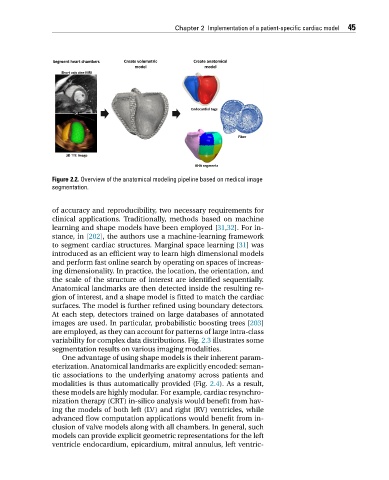
Figure 2.2. Overview of the anatomical modeling pipeline based on medical image
segmentation.
of accuracy and reproducibility, two necessary requirements for
clinical applications. Traditionally, methods based on machine
learning and shape models have been employed [31,32]. For in-
stance, in [202], the authors use a machine-learning framework
to segment cardiac structures. Marginal space learning [31]was
introduced as an efficient way to learn high dimensional models
and perform fast online search by operating on spaces of increas-
ing dimensionality. In practice, the location, the orientation, and
the scale of the structure of interest are identified sequentially.
Anatomical landmarks are then detected inside the resulting re-
gion of interest, and a shape model is fitted to match the cardiac
surfaces. The model is further refined using boundary detectors.
At each step, detectors trained on large databases of annotated
images are used. In particular, probabilistic boosting trees [203]
are employed, as they can account for patterns of large intra-class
variability for complex data distributions. Fig. 2.3 illustrates some
segmentation results on various imaging modalities.
One advantage of using shape models is their inherent param-
eterization. Anatomical landmarks are explicitly encoded: seman-
tic associations to the underlying anatomy across patients and
modalities is thus automatically provided (Fig. 2.4). As a result,
these models are highly modular. For example, cardiac resynchro-
nization therapy (CRT) in-silico analysis would benefit from hav-
ing the models of both left (LV) and right (RV) ventricles, while
advanced flow computation applications would benefit from in-
clusion of valve models along with all chambers. In general, such
models can provide explicit geometric representations for the left
ventricle endocardium, epicardium, mitral annulus, left ventric-