Page 97 - Chiral Separation Techniques
P. 97
3.7 Combinatorial Libraries of Selectors for HPLC 73
The experiments of the initial study were performed on a 5 g scale. Although fully
feasible, this quantity of functionalized beads required the use of very substantial
amounts of reagents and solvents for both the preparation and the chromatographic
testing. Therefore, the same group later developed a microscale methodology for
screening with only 50 mg of the CSP [85]. Their approach assumed that if an effi-
cient CSP is placed in a solution of a racemate, it should adsorb preferentially only
one of the enantiomers, thus depleting it from the solution. To confirm this assump-
tion, they placed a few milligrams of the beads in a vial and added a dilute solution
of a less than equimolar amount of the target racemate. After equilibration, a sample
of the supernatant liquid was injected into a commercial chiral HPLC column and
the peak areas were determined for both separated enantiomers to calculate the
selectivity. If the areas were equal, the CSP did not exhibit any selectivity, while any
change in the amount of an enantiomer represented some level of selectivity.
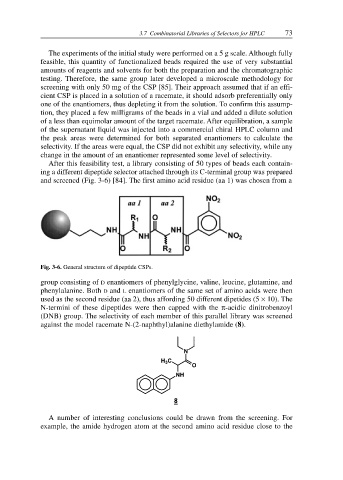
After this feasibility test, a library consisting of 50 types of beads each contain-
ing a different dipeptide selector attached through its C-terminal group was prepared
and screened (Fig. 3-6) [84]. The first amino acid residue (aa 1) was chosen from a
Fig. 3-6. General structure of dipeptide CSPs.
group consisting of D enantiomers of phenylglycine, valine, leucine, glutamine, and
phenylalanine. Both D and L enantiomers of the same set of amino acids were then
used as the second residue (aa 2), thus affording 50 different dipetides (5 × 10). The
N-termini of these dipeptides were then capped with the π-acidic dinitrobenzoyl
(DNB) group. The selectivity of each member of this parallel library was screened
against the model racemate N-(2-naphthyl)alanine diethylamide (8).
A number of interesting conclusions could be drawn from the screening. For
example, the amide hydrogen atom at the second amino acid residue close to the