Page 90 - Computational Modeling in Biomedical Engineering and Medical Physics
P. 90
78 Computational Modeling in Biomedical Engineering and Medical Physics
Before the manual postprocessing of the segmented mask, a flood-filling algorithm
(Fathi and Hiltner, 1999) should be used. This will check for discontinuities in the
mask and will eliminate the disconnected regions (islands). Also a cavity removal tool
(Chris and Garland, 1990) should repair the mask, filling in the cavities, once applied.
The source image quality, strongly related to the overlapping artifacts and noise,
affects the ROI reconstruction process. Even a well-optimized segmentation algorithm
will provide a preliminary 3D model of the ROI with a set of additional unwanted
features (Fig. 3.6D). These should be carefully removed, without accidentally touching
the mask of the ROI. For example, smoothing filters (Kuan et al., 1985) and noise
removal tools (Rudin et al., 1992), selected and configured correctly, could help at
generating better masks by the user-controlled removal of the undesirable regions
(Fig. 3.6E). The smoothing filters attenuates the images noise level and evens the sharp
contours and edges of the ROI, while the noise removal algorithms are mainly con-
cerned with improving the images background.
The 3D-smoothed mask (Fig. 3.6E) is made of both blood and bone tissue. Thus the
latter needs to be removed to create a 3D mask of the arterial network. This process is
manual and consists of carefully selecting and deleting thebonetissuestraightfromthe 2D
mask as depicted in the source images (Fig. 3.6A C) or from the 3D solid model gener-
ated by the segmentation algorithm (Fig. 3.6E). Although the 3D elimination process of
thebonetissueiseasierbecause it brings the advantage of a more precise and effortless
selection of the unwanted regions, it needs enough hardware resources, especially 3D gra-
phics, to work well. This aspect becomes significant when dealing with complex models,
comprised of several masks, for different types of organs or tissues.
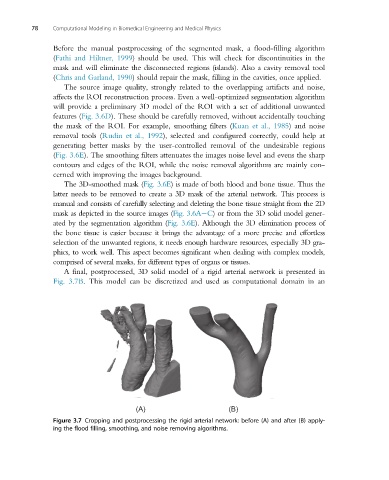
A final, postprocessed, 3D solid model of a rigid arterial network is presented in
Fig. 3.7B. This model can be discretized and used as computational domain in an
(A) (B)
Figure 3.7 Cropping and postprocessing the rigid arterial network: before (A) and after (B) apply-
ing the flood filling, smoothing, and noise removing algorithms.