Page 299 - Thermodynamics of Biochemical Reactions
P. 299
Thermodynamics of Biochemical Reactions at Specified pH 299
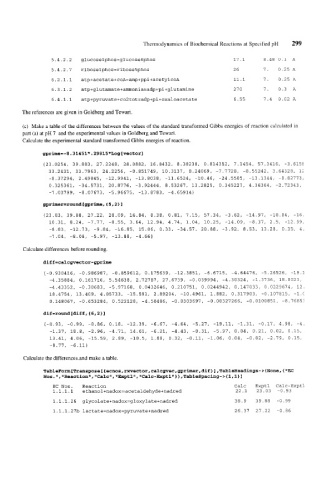
5.4.2.2 glucoselphos=glucose6phos 17.1 8.48 U.1 A
5.4.2.7 riboselphos=ribose5phos 26 7. 0.25 A
6.2.1.1 atp+acetate+coA=amp+ppi+acetylcoA 11.1 7. 0.25 A
6.3.1.2 atp+glutamate+ammonia=adp+pi+glutamine 27 0 7. 0.3 A
6.4.1.1 atp+pyruvate+co2tot=adp+pi+oxaloacetate 6.55 7.4 0.02 A
The references are given in Goldberg and Tewari.
(c) Make a table of the differences between the values of the standard transformed Gibbs energies of reaction calculated in
part (a) at pH 7 and the experimental values in Goldberg and Tewari.
Calculate the experimental standard transformed Gibbs energies of reaction.
gprime=-8.31451*.29815*Log[vectorl
{23.0254, 39.883, 27.2248, 28.0882, 16.8432, 8.38238, 0.814352, 7.1454, 57.3416, -3.615E
33.2431, 33.7963, 24.2256, -0.851749, 10.3137, 8.24069, -7.7728, -8.55242, 3.64328, 1;
-8.37294, 2.49845, -12.9941, -13.8038, -11.6524, -10.46, -24.5505, -13.1344, -8.82773,
0.325361, -34.5731, 20.8776, -3.92444, 8.53267, 13.2825, 0.345227, 4.36364, -2.72343,
-7.03799, -8.07673, -5.96675, -13.8783, -4.65914)
gprimer=round [gprime, { 5,2 1 1
(23.03, 39.88, 27.22, 28.09, 16.84, 8.38, 0.81, 7.15, 57.34, -3.62, -14.97, -18.84, -16.
10.31, 8.24, -7.77, -8.55, 3.64, 12.94, 4.74, 1.04, 10.25, -14.09, -8.37, 2.5, -12.99,
-8.83, -12.73, -9.84, -16.85, 15.06, 0.33, -34.57, 20.88, -3.92, 8.53, 13.28, 0.35, 4.
-7.04, -8.08, -5.97, -13.88, -4.66)
Calculate differences before rounding.
diff=calcgvector-gprime
{-0.930416, -0.986987, -0.859612, 0.175639, -12.3851, -6.6715, -4.64476, -5.26526, -19.1
-4.35884, 0.161716, 5.54638, 2.72787, 27.6739, -0.039994, -4.30324, -1.3736, 18.8023,
-4.43352, -0.30683, -5.97168, 0.0432646, 0.210751, 0.0244942, 0.147033, 0.0229674, 12.
18.4754, 13.409, 4.05733, -15.591, 2.89204, -10.4961, 1.882, 0.317903, -0.107815, -1.c
0.148067, -0.653284, 0.522128, -4.50486, -0.0303697, -0.00327266, -0.0100851, -8.7685:
dif =round [dif f, { 6,2 11
{-0.93, -0.99, -0.86, 0.18, -12.39, -6.67, -4.64, -5.27, -19.11, -1.31, -0.17, 4.98, -4.
-1.37, 18.8, -2.96, -4.71, 14.61, -6.21, -4.43, -0.31, -5.97, 0.04, 0.21, 0.02, 0.15,
13.41, 4.06, -15.59, 2.89, -10.5, 1.88, 0.32, -0.11, -1.06, 0.04, -0.02, -2.79, 0.15,
-8.77, -6.11}
Calculate the differences.and make a table.
TableForm[Transpose[{ecnos,rxvector,calcgvec,gprimer,dif}],TableHead~ngs-~iNone,i"EC
Nos.", 18Reaction", "Calc", Wxptl1I, 'Calc-Exptl") 1 ,TableSpacing->{l, 111
EC Nos. Reaction Calc Exptl Calc-Exptl
1.1.1.1 ethanol+nadox==acetaldehyde+nadred 22.1 23.03 -0.93
1.1.1.26 glycolate+nadox=gloxylate+nadred 38.9 39.88 -0.99
1.1.1.27b lactate+nadox=pyruvate+nadred 26.37 27.22 -0.86