Page 499 -
P. 499
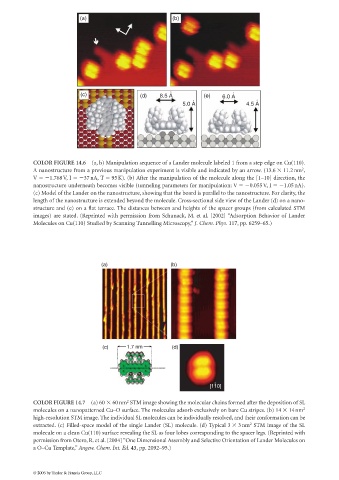
(a) (b)
(c) (d) 8.5 A ° (e) 6.0 A °
° °
5.0 A 4.5 A
COLOR FIGURE 14.6 (a, b) Manipulation sequence of a Lander molecule labeled 1 from a step edge on Cu(110).
2
A nanostructure from a previous manipulation experiment is visible and indicated by an arrow. (13.6 11.2 nm ,
V 1.768V, I 37nA, T 95K). (b) After the manipulation of the molecule along the [1–10] direction, the
nanostructure underneath becomes visible (tunneling parameters for manipulation: V 0.055 V, I 1.05 nA).
(c) Model of the Lander on the nanostructure, showing that the board is parallel to the nanostructure. For clarity, the
length of the nanostructure is extended beyond the molecule. Cross-sectional side view of the Lander (d) on a nano-
structure and (e) on a flat terrace. The distances between and heights of the spacer groups (from calculated STM
images) are stated. (Reprinted with permission from Schunack, M. et al. [2002] “Adsorption Behavior of Lander
Molecules on Cu(110) Studied by Scanning Tunnelling Microscopy,” J. Chem. Phys. 117, pp. 6259–65.)
(a) (b)
(c) 1.7 nm (d)
-
[110]
2
COLOR FIGURE 14.7 (a) 60 60nm STM image showing the molecular chains formed after the deposition of SL
molecules on a nanopatterned Cu–O surface. The molecules adsorb exclusively on bare Cu stripes. (b) 14 14 nm 2
high-resolution STM image. The individual SL molecules can be individually resolved, and their conformation can be
extracted. (c) Filled-space model of the single Lander (SL) molecule. (d) Typical 3 3nm STM image of the SL
2
molecule on a clean Cu(110) surface revealing the SL as four lobes corresponding to the spacer legs. (Reprinted with
permission from Otero, R. et al. [2004] “One Dimensional Assembly and Selective Orientation of Lander Molecules on
a O–Cu Template,” Angew. Chem. Int. Ed. 43, pp. 2092–95.)
© 2006 by Taylor & Francis Group, LLC