Page 424 - Numerical Methods for Chemical Engineering
P. 424
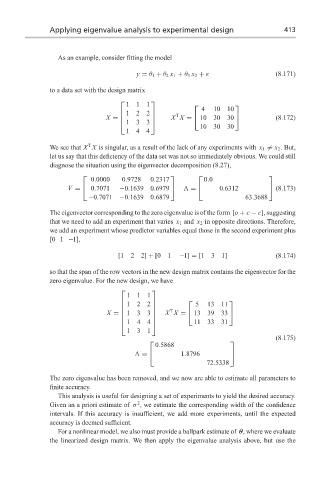
Applying eigenvalue analysis to experimental design 413
As an example, consider fitting the model
y = θ 1 + θ 2 x 1 + θ 3 x 2 + ε (8.171)
to a data set with the design matrix
111
4 10 10
122
T
X = X X = 10 30 30 (8.172)
133
10 30 30
144
T
We see that X X is singular, as a result of the lack of any experiments with x 1 = x 2 . But,
let us say that this deficiency of the data set was not so immediately obvious. We could still
diagnose the situation using the eigenvector decomposition (8.27),
0.0000 0.9728 0.2317 0.0
V = 0.7071 −0.1639 0.6979 = 0.6312 (8.173)
−0.7071 −0.1639 0.6879 63.3688
The eigenvector corresponding to the zero eigenvalue is of the form [o + c − c], suggesting
that we need to add an experiment that varies x 1 and x 2 in opposite directions. Therefore,
we add an experiment whose predictor variables equal those in the second experiment plus
[0 1 −1],
[122] + [01 −1] = [131] (8.174)
so that the span of the row vectors in the new design matrix contains the eigenvector for the
zero eigenvalue. For the new design, we have
111
5 13 11
122
T
X X = 13 39 33
X = 133
11 33 31
144
131
(8.175)
0.5868
= 1.8796
72.5338
The zero eigenvalue has been removed, and we now are able to estimate all parameters to
finite accuracy.
This analysis is useful for designing a set of experiments to yield the desired accuracy.
2
Given an a priori estimate of σ , we estimate the corresponding width of the confidence
intervals. If this accuracy is insufficient, we add more experiments, until the expected
accuracy is deemed sufficient.
For a nonlinear model, we also must provide a ballpark estimate of θ, where we evaluate
the linearized design matrix. We then apply the eigenvalue analysis above, but use the